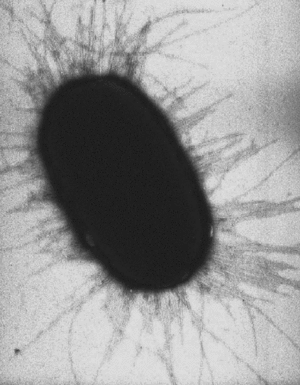
Pilus
A pilus (Latin for 'hair'; plural : pili) is a hairlike protein structure on the surface of a cell, especially Gram-negative bacteria.
A pilus is typically 7 nm in diameter , and several hundred pili can extend from the surface of a bacterial cell [1] (see image to right).
A fimbria (Latin meaning fringe; plural: fimbriae) is a short pilus. They are used for attachment of the cell to a surface or other cells, andare either located at the poles of a cell, or are evenly spread over its entire surface. In some cases, mutant pathogenic bacteria that lack fimbriae cannot adhere to their usual target host cell surfaces, and thus cannot cause disease.
The terms pili and fimbriae are often used interchangeably. Numerous different types of pili have been characterized, and various forms of these appendages are involved in diverse activities of bacteria. These include bacterial cell aggregation, adhesion to surfaces of host cells such as, in the case of gut bacteria, the linings of the intestine, adhesion to other microbial cells in biofilms, gene and protein injection into other cells, DNA uptake by naturally transformable bacteria, and virulence attributes of pathogenic bacteria.
Pili are distinctly different from bacterial flagella; unlike flagella they grow from the inside of the cell outward, and not from the tip of the fiber.
Bacterial fimbriae can contain lectin proteins generally at their tips. These lectins are nused for adherance to target cells such as intestinal cells in the gut because they can recognize oligosaccharide units such as mannose on the surface of these target cells. These lectins are one of the many types of adhesin protein produced by bacteria. However, all types of pili are primarily composed of oligomeric pilin proteins which form the pilus rod structure.
Some forms of pili are encoded by plasmids, for instance fertility factor F encodes F-pili (sex-pili) which are present at only 2 to 4 per cell. F-pili are involved in bacterial conjugation and connect the bacterium to another bacterium and build a bridge between the cytoplasms of the cells which becomes the channel for one way transfer of a single-strand of DNA and certain protein molecules. This transfer enables the mobilization of plasmids between bacteria. An exchanged plasmid can add new functions to a bacterium, e.g., an antibiotic resistance.
Some bacterial viruses (bacteriophages) attach to receptors on sex pili at the start of their reproductive cycle.
Type I pili
Type I pili are widely distributed on enteric bacteria and their structure and assembly is well characterized. They are composed of several different protein components including FimH adhesin that is part of the short thin fibrillar pilus tip, and FimA which makes up the thicker 7 nm thick long rod of the pilus. FimA binds to mannose sugars present on a variety of different host cell surface structures and can be considered to be a lectin [2].
The assembly of the Type I pilus occurs in the periplasm and involves the "chaperone/usher" protein folding and delivery system.
Sex pili
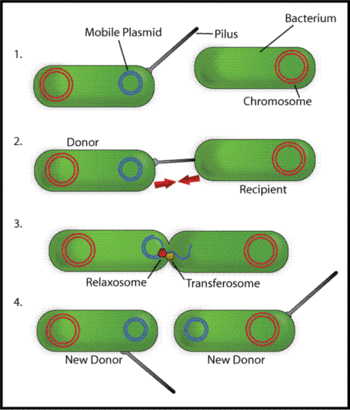
The pili encoded by F plasmid in Escherichia coli are involved in formation of cell aggregates as a prelude to gene transfer, called conjugation, from the F-plamid containing cells to a recipient (see figure) [3]. The F-pilus itself has an elaborate structure comprising several different proteins, encoded by numerous genes on the F-plasmid that are involved in pilus formation and DNA transfer, including genes traA, E, K, B, V, W, U, F and G, [4].
Adhesion of bacterial cells has an important survival role in their survival as micro-colonies - called biofilms - on solid surfaces in the natural environment, and F-pili determine the final shapes of the structures seen in mature surface biofilms formed by Eschericha coli bacteria, as mutants affected in the plasmid specified F-pili form a biofilm of a different structure[5].
The pilus allows for the transfer of bacterial DNA from the F-piliated (donor) bacteria to the recipient bacteria by conjugation, and similar transfer functions are carried by many different plasmids such as the R-plasmids that confer antibiotic resistance. Through this mechanism of conjugation based gene transfer, advantageous genetic traits can be widely disseminated amongst populations of bacteria.
Although not all bacteria have the ability to create sex pili, sex pili can form mating channels between bacteria of different species, allow mobilisation of other plasmids such as ColE1 that themselves possess no conjugation ability, and even promote DNA transfer between bacteria and eukaryotic cells such as those of yeasts and plants. The VirB protein of crown-gall bacterium Agrobacterium tumefaciens is a pilin like protein similar to protein TraA of E. coli F-plasmid. VirB is involved in the injection of T-DNA into plant cells by the crown-gall bacterium[6].
Type 4 pili
Type 4 pili are thin, flexible , 6- to 7-nm fibers displayed by a wide variety of gram-negative bacterial species and members of this class are recognisable because of amino-acid sequence similarities. They have a distinctive method of assembly involving proteolytic processing and N-methylation of the pilin precursor, and they are not implicated in conjugation. Components of the pilus are also involved in natural DNA uptake (transformation) in Neisseria bacteria. Type 4 pili are primarily involved in adhesion to surfaces and as virulence determinants in pathogenic bacteria. In some species they cause twitching motility, for instance in Pseudomonas aeruginosa, Myxococcus and Neisseria bacteria [7].
Twitching motility is well understood in the pathogenic diplococcus Neisseria gonorrhoeae which has has been shown to move by first extending its Type 4 pili so that the external termini of the pili adhere to a solid substrate, and subsequently using pilus retraction to pull the bacteria forward, not unlike a grappling hook[8] .
Pili of pathogens
Type 4 pili are produced by the enteric pathogenic bacteria Vibrio cholerae, enteropathogenic Escherichia coli, and enterotoxigenic E. coli. Bundle forming pili (BfpA) of enteropathogenic Escherichia coli are Type IV pili that are necessary for adhesion to epithelial cells. Type 4 pili are virulence factors of Neisseria meningitidis and N. gonorrhoeae bacteria. Many bacteria producing Type 4 pili also produce other adhesion factors [9] [10].
Illustrations of these pilus-target interactions, and of the remarkable design features of these powerful bacterial grappling devices, are found in the PLoS Biology article that follows.
Bacterial fimbriae designed to stay with the flow
Liza Gross
The human digestive system houses a diverse colony of beneficial bacteria, but one species—E. coli—can wreak havoc when it colonizes mucous membranes that normally exist unmolested (for example, in the urinary tract). To latch on to cells and establish infection, E. coli uses fimbriae—long, hairlike organelles that project from the bacterium’s surface. Fimbriae consist of interlinking subunits of a single protein called pilin that forms a rigid, coiled helix-shaped rod. Sticky proteins called adhesins cap the tip of the rod and bind to carbohydrate receptors on their host, thus securing bacteria on the host cells as extracellular fluids swirl around them.
A previous study led by Evgeni Sokurenko and Viola Vogel investigated the most common type of E. coli fimbriae. The sticky protein at the tip of these fimbriae is called FimH and binds to a carbohydrate called mannose. They showed that powerful drag forces created by the extracellular fluids don’t carry the bound bacteria away, as one might expect, but instead strengthen their adhesion to their host. The researchers attributed this increased binding to a biphasic “catch bond” mechanism whereby increased drag forces cause the FimH at the tip of the fimbria to switch from a form that binds mannose weakly to a form that binds strongly. Because of this, the bacteria bind best at an optimal force that is high enough to switch FimH to strong binding but not so high that it breaks the strong FimH–mannose bond.
And now, in a new study, the same group of researchers (including first author Manu Forero) set out to determine whether the coiled rod structure of fimbriae affects how the sticky FimH at the tip binds. It had been assumed that fimbrial rods play a largely static structural role, either by extending the tip adhesins’ reach or by resisting electrostatic repulsive forces between bacteria and cell surfaces. But Forero et al. show that the rods function more dynamically, using their mechanical properties to help stabilize the FimH–mannose bond against a turbulent background.
Fimbriae-mediated adhesion was investigated with an atomic force microscope, which uses a cantilever to apply (and measure) forces between its tip and the sample under investigation. Forero et al. outfitted the cantilever tip with mannose, and then used this to touch a fimbriated E. coli cell that was affixed to a glass surface. After mannose bound to the fimbrial FimH, the cantilever retreated from the bacterium at a constant velocity. The researchers determined that, instead of the FimH–mannose bond breaking, the fimbriae stretched out far beyond their original length.
One reason that fimbriae extend could be that the individual pilin subunits of the fimbrial rod are uncoiling. The researchers tested this hypothesis by applying a constant force between the cantilever and fimbria—under which fimbrial length changes slowly. They observed that stepwise jumps in distance corresponded to the expected length of individual subunits unwinding from the coiled shaft one at a time. Thus, the researchers concluded, fimbrial elongation proceeds as subunits uncoil one after another. This was also supported by a mathematical model developed by the researchers to quantify the biophysical forces governing the dynamics of fimbrial uncoiling.
Forero et al. also detected that, after uncoiling at increasing force, the stretched fimbriae re-coil if the pulling force drops. Importantly, while fimbrial uncoiling under high force decreases the tension within the rod, re-coiling under low force increases the tension. Thus, the tensile force within the rod stays within some intermediate level when fimbrial length is stable.
The researchers found that the intermediate force range corresponds to the force level where the FimH–mannose bonds last longest. Lower, coiling forces are too weak to switch bonds to a long-lived state before breaking, and higher, uncoiling forces exceed the catch-bond threshold, shortening the life of the bond. Because E. coli living in the gut or other mucosal surfaces experience constantly changing flow rates and forces, these adjustments should enhance fimbrial attachment under a diverse range of fluid conditions. The correspondence of forces suggests that the mechanical properties of the fimbrial rod and the FimH–mannose complex co-evolved to optimize adhesive stability in fluids.
- Citation: Gross L (2006) Bacterial Fimbriae Designed to Stay with the Flow. PLoS Biol 4(9): e314 DOI: 10.1371/journal.pbio.0040314 Published: August 29, 2006
- Copyright: © 2006 Public Library of Science. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
Citations
- ↑ Brinton, Charles C., Jr.; Gemski, Peter., Jr.; Carnahan, Judith. A NEW TYPE OF BACTERIAL PILUS GENETICALLY CONTROLLED BY THE FERTILITY FACTOR OF E. COLI K 12 AND ITS ROLE IN CHROMOSOME TRANSFER. Proc Natl Acad Sci U S A. 1964 Sep;52(3):776–783.
- ↑ CH Jones, JS Pinkner, R Roth, J Heuser, AV Nicholes, SN Abraham and SJ Hultgren (1995) FimH Adhesin of Type 1 pili is Assembled into a Fibrillar tip Structure in the Enterobacteriaceae Proceedings of the National Academy of Sciences, Vol 92, 2081-2085
- ↑ Achtman, A. (1975) Mating aggregates in Escherichia coli conjugation . J Bacteriol. August; 123(2): 505–515.
- ↑ Frost, L. S. Ippen-Ihler, K., and Skurray, R. A. (1994). Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiological Reviews 58, p162-210.
- ↑ Reisner, Andreas, Haagensen, Janus A. J. , Schembri, Mark A., Zechner, Ellen L., and Molin, Søren (2003) Development and maturation of Escherichia coli K-12 biofilms Molecular Microbiology 48 (4), pages 933–946
- ↑ Jones AL, Lai EM, Shirasu K, Kado CI (1996) VirB2 is a processed pilin-like protein encoded by the Agrobacterium tumefaciens Ti plasmid.J Bacteriol. 1996 Oct;178(19):5706-11.
- ↑ Burrows LL.(2005) Weapons of mass retraction. Mol Microbiol. 2005 Aug;57(4):878-88.
- ↑ Merz AJ, So M, Sheetz MP.(2000) Pilus retraction powers bacterial twitching motility. Nature. 2000 Sep 7;407(6800):98-102.
- ↑ Marsh JW, Taylor RK. (1999) Genetic and transcriptional analyses of the Vibrio cholerae mannose-sensitive hemagglutinin type 4 pilus gene locus.
- ↑ J Bacteriol. 1999 Feb;181(4):1110-7.Strom, M. S., and S. Lory. (1993). Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47 pages 565-596.
Further reading
- Todar's Online Textbook of Bacteriology MECHANISMS OF BACTERIAL PATHOGENICITY: COLONIZATION AND INVASION
- CH Jones, JS Pinkner, R Roth, J Heuser, AV Nicholes, SN Abraham and SJ Hultgren (1995) FimH Adhesin of Type 1 pili is Assembled into a Fibrillar tip Structure in the Enterobacteriaceae Proceedings of the National Academy of Sciences, Vol 92, pages 2081-2085
See also