Mycobacterium tuberculosis: Difference between revisions
imported>Meg Taylor No edit summary |
mNo edit summary |
||
| Line 104: | Line 104: | ||
==References== | ==References== | ||
{{reflist|2}} | {{reflist|2}}[[Category:Suggestion Bot Tag]] | ||
Latest revision as of 06:01, 22 September 2024
 | ||||||||||||||
| Scientific classification | ||||||||||||||
| ||||||||||||||
| Binomial name | ||||||||||||||
| Mycobacterium tuberculosis |
Description and significance
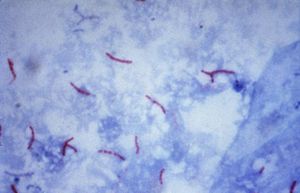
Mycobacterium tuberculosis was first isolated and described in 1882 by Dr. Robert Koch, who later went on to win the Nobel Prize in 1905 for his discovery. Also known as, "Tubercle Bacillus", and Koch's bacillus, M. Tuberculosis is a non-motile, rod shaped bacterium, and the primary cause for the widespread disease tuberculosis.
The World Health Organization estimates that one-third of the world's population is infected with M. Tuberculosis, and that someone new is infected every second. [1] With the emergence of drug resistant strains, both multi drug resistant (MDR) and extensively drug resistant (XDR), controlling the spread of tuberculosis has become a major concern. Since 2006, the WHO's primary strategy in its Stop TB Strategy has been the use of DOTS (Directly Observed Treatment, Short-course). It is estimated to be the most cost effective strategy, only costing $3-$7 to gain a year of health. [2]
Genome structure
The genome for two strains of M. Tuberculosis have been sequenced completely. In 1998, the Sanger Institute sequenced the common laboratory strain, H37rv. At the same time, strain CD1551 was sequenced by The Institute for Genomic Research (TIGR) using the whole genome random sequencing method (shotgun sequencing). The CDC-1551 strain has been shown to be more virulent than the H37rv strain. By comparing both genomes, key information can be obtained about the virulence, survivability (drug resistance), and evolution of M. Tuberculosis. [3][4]
Chart from "TIGR"summarizing the CDC-1551 strain
| DNA Molecule Summary | ||
| Total Number of all DNA molecules: | 1 | 100.00% |
| Total Size of all DNA molecules: | 4403837 bp | 100.00% |
| Number of Primary Annotation coding bases: | 4069477 bp | 92.40% |
| Number of G+C bases: | 2889216 bp | 65.60% |
| Primary Annotation Summary | ||
| Total genes: | 4294 | 100.00% |
| Protein coding genes: | 4246 | 98.88% |
| Genes assigned a role category: | 2317 | 54.56% |
| Genes not assigned a role category: | 2 | 0.04% |
| Conserved hypothetical genes: | 655 | 15.42% |
| Hypothetical genes: | 1272 | 29.95% |
| tRNA genes: | 45 | 1.04% |
| rRNA genes: | 3 | 0.06% |
Cell structure and metabolism
M. Tuberculosis is a non-motile, rod shaped bacterium roughly 2-4 um in length and 0.2-0.5 um in width. It's an obligate aerobe and neither Gram-positive or Gram-negative. Although it has a peptidoglycan layer, M. Tuberculosis does not retain the crystal violet dye. To solve this issue, acid-fast staining is used (specifically Ziehl-Neelsan Stain). The peptidoglycan layer is of particular interest. It is composed mostly of lipids (60%). The high lipid percentage (lipids: Mycolic Acid, Cord Factor, Wax-D) is the likely cause of the high survivability of M. Tuberculosis. Factors include: impermeability to stains; resistance to antibiotics, osmotic lysis, acidic and alkaline compounds, and protection from oxygen free radicals. other factors that add to the survivability of M. Tuberculosis include the ability to grow intracellularly within macrophages (exact reason is unknown), and it's slow rate of growth compared to other bacteria (about 15-20 hours). The slow rate of growth may deter it's detection by the immune system.[5]
Ecology
M. Tuberculosis generally resides in the lungs where oxygen is more readily available. However, if the bacteria enters the blood stream, or lymphatic system, it can spread throughout the body and infect various other organs (brain, kidney, liver, etc.) When this happens, it is known as miliary tuberculosis.
M. Tuberculosis has an increased capability to survive outside our system due to its highly lipid based peptidoglycan layer. Within our system, M. Tuberculosis are consumed by macrophages, however lysosome-phagosome fusion is prevented, and M. TB is able to grow inside the macrophages.[6][7]
Pathology
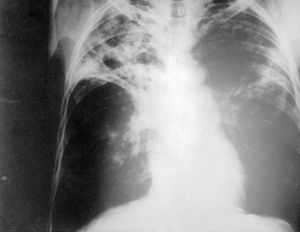
M. Tuberculosis is the major cause of tuberculosis. Transmission of M. TB is by means of droplet nuclei which contain the bacteria. Coughing (a common symptom of tuberculosis), talking, singing, and definitely sneezing are methods of transmission. Due to the small size of the droplets, they are able to linger in the air for an extended amount of time. After inhalation of the bacteria, it will make its way to your lungs, where macrophages will consume them. However, M. Tuberculosis prevents lysome-phagosome fusion and as a result is not destroyed. Continually growing in the macrophages, a cell mediated immune response will occur. The body will attempt to quarantine the bacteria (use of macrophages and T cells), as a result, Tuberculosis nodules are formed, or more generally granulomas. However, due to the increased survivability, and persistence of M. TB, in some cases it may be able to continue to grow. Eventually spreading from the alveoli to the bronchi and then throughout the lungs. In rare cases, M. tuberculosis may be able to enter the blood stream, or lymphatic systems, the resulting disease is know as Miliary Tuberculosis. Miliary Tuberculosis is estimated to only occur in 1.5% of all TB patients. [8]
Current Research
Currently the only vaccine for tuberculosis is the bacille Calmette-Guerin (BCG) vaccine which comes from an attenuated strand of mycobacterium bovis. The BCG vaccine is not used widely in many countries, including the United States, due to the fact that it causes a false positive result with the tuberculin skin test (TST), or sometimes simply known as a PPD (purified protein derivative). There has been research ongoing that has discovered that two proteins, namely narK2 (Rv1737c) and narX (Rv1736c), may be used to differentiate between the BCG vaccine, and latent tuberculosis. [9]
Other vaccines are undergoing clinical trials, and one of the main reasons allowing this to happen is the complete sequencing of the genome. With the genome sequenced, it is possible for researches to study enzymes that they previously could not due to the fact that the expression of the enzyme was low. One such case is the study of lipolytic enzymes that degrade internal and external lipids. The research team theorizes that this lipolytic enzyme may be correlated to the virulence of mycobacterium tuberculosis. [10]
As often preached, prevention is one of the keys to controlling a disease. Obviously however, treatment for those diseased are vital as well. Although there are numerous ways to treat M. tuberculosis, there has been a surge in strains that are resistant to our current drugs. Both multi-drug resistance, and extensively drug resistance is on the rise worldwide. By understanding the phenotypical/genotypical differences between strains with resistance and those without, it may be possible to control the surge of drug resistant strains. [11]
References
- ↑ http://www.who.int/mediacentre/factsheets/fs104/en/index.html
- ↑ http://www.tbalert.org/worldwide/DOTS.php
- ↑ http://cmr.tigr.org/CMR/org_gmt.shtml
- ↑ http://www.biohealthbase.org/GSearch/aboutMycobacterium.do?decorator=Mycobacterium
- ↑ http://www.textbookofbacteriology.net/tuberculosis.html
- ↑ http://www.biohealthbase.org/GSearch/aboutMycobacterium.do?decorator=Mycobacterium
- ↑ http://www.textbookofbacteriology.net/tuberculosis.html
- ↑ http://www.emedicine.com/med/TOPIC1476.HTM
- ↑ http://www.ncbi.nlm.nih.gov/pubmed/18362135?ordinalpos=6&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum%7C
- ↑ http://www.ncbi.nlm.nih.gov/pubmed/18309478?ordinalpos=19&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum%7C
- ↑ http://jac.oxfordjournals.org/cgi/content/full/59/5/866