Horizontal gene transfer in prokaryotes: Difference between revisions
imported>David Tribe |
mNo edit summary |
||
| (61 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{subpages}} | |||
{{main|Horizontal gene transfer}} | {{main|Horizontal gene transfer}} | ||
[[Image:J_Lederberg.jpg|frame|[[Joshua Lederberg]] in the 1960s (NLM). His 1946 discovery of mating in ''[[Escherichia coli]]'' was heralded by [[Salvador Luria]] as likely to be "among the most fundamental advances in the whole history of [[bacteriology|bacteriological]] science".<ref>Luria, S. E. (1947). Recent advances in bacterial genetics. Bacteriological Reviews 11, page 1.</ref>. For this work he was awarded the [[Nobel Prize in Physiology or Medicine]] for 1958. The bacterial mating mechanism, called '''conjugation''', a major mechanism for horizontal gene transfer, is now known to have to have [[evolution|evolved]] to perform a wide range of biological roles for injection of both [[DNA]] and [[protein]] into diverse target [[cell (biology)|cells]], including [[bacteria]], [[yeast]]s, [[plant (organism)|plants]] and [[protist]]s.]] | |||
In the years 1987-1996 in [[Vietnam]] there was a medically alarming appearance of resistance to the [[antibiotics|antibiotic]] [[chloramphenicol]] in dangerous [[meningococcus|meningococci]] [[bacteria]] that cause infectious [[meningitis]], hampering the treatment of the [[disease]] in that country. Subsequent laboratory investigations revealed that the [[gene]] conferring chloramphenicol resistance on pathogenic meningococcus bacteria was identical to a previously identified [[transposon|mobile gene]] named ''Tn4451'' found in the completely different bacteria known as ''Clostridium perfringens''.<ref>Galimand M, Gerbaud G, Guibourdenche M, Riou JY, Courvalin P.(1998) High-level chloramphenicol resistance in ''Neisseria meningitidis''. N Engl J Med. 1998 Sep 24;339(13):868-74. Erratum in: Engl J Med 1999 Mar 11;340(10):824.Comment in: N Engl J Med. 1998 Sep 24;339(13):917-8.</ref> Meningococci are [[Gram negative]] and [[aerobic]] [[microbe]]s, while Clostridium is Gram positive and [[anaerobic]]- about as different from one-another as bacteria can be. | |||
''' | This episode is but one illustration of the extensive movement of genes that takes place between distantly related microbes. This process has the technical name of '''[[horizontal gene transfer]]''' (HGT) and is defined as movement of genes between different [[species]], or across broad [[taxonomy|taxonomic]] categories. | ||
As indicated by the opening example, HGT is common among both the two major taxonomic subdivisions of bacteria-like organisms - ''[[bacterium|Bacteria]]'' and ''[[Archaea]]'' - and occurs even between very distantly-related microbes. Both the Bacteria and Archaea taxonomic Domains of [[microorganism]] lack distinct nuclear cellular compartments for their DNA, and are thus classified as '''[[Prokaryote|prokaryote]]s''', meaning they possess a primitive nucleus, in contrast to [[Eukaryote|eukaryote]] cells which possess well-formed nuclei. (HGT also occurs in eukaryotic [[protist]] microorganisms, and in both pathogenic and non-pathogenic microorganisms, so overall is a major feature of natural microbial evolution.) | |||
Detailed and comprehensive DNA sequence analysis of many prokaryotic cell genomes has shown that these genomes generally consist of a conserved backbone of mainly ''housekeeping'' genes that is subject to interruption by ''islands'' of DNA. These DNA islands can change relatively frequently during prokaryote evolution due to insertion, deletion and replacement events involving by foreign DNA.<ref> Levin BR Bergstrom CT (2000) [http://books.nap.edu/openbook.php?record_id=9766&page=R1 Bacteria are Different: Observations, Interpretations, Speculations, and Opinions About the Mechanisms of Adaptive Evolution in Prokaryotes], Chapter 7 in Francisco J. Ayala, Walter M. Fitch, and Michael T. Clegg, Editors, Variation and Evolution in Plants and Microorganisms, Towards a New Synthesis 50 years after Stebbing. National Academy of Sciences, Washington DC. also available as Levin BR, Bergstrom CT.(2000) [http://www.pnas.org/cgi/content/full/97/13/6981 Bacteria are different: observations, interpretations, speculations, and opinions about the mechanisms of adaptive evolution in prokaryotes] .Proc. Natl. Acad. Sci. U.S.A. 2000 Jun 20;97(13):6981-5. PMID 10860960 </ref> | |||
Studies of antibiotic resistance genes provide convincing evidence for wide transmissibility of genes between taxonomically diverse microbial species. | |||
==Main mechanisms of gene transfer in prokaryotes== | ==Main mechanisms of gene transfer in prokaryotes== | ||
HGT between distant microbial [[taxon|taxa]] is mediated by [[bacteriophages]], [[plasmids]], [[transposons]], and specialized cellular DNA uptake mechanisms. These mechanisms usually categorized with the following commonly used technical terms: | |||
* '''Bacterial [[Transformation (genetics)|transformation]]''' , the genetic alteration of a [[cell (biology)|cell]] resulting from the introduction, uptake and [[expression (genetics)|expression]] of foreign genetic material, generally [[DNA]] (see this [http://www.learner.org/channel/courses/biology/archive/animations/hires/a_infect3_h.html animation with audio explanation]). This process is relatively common in certain naturally transformable bacteria, such as the soil bacterium ''[[Bacillus subtilis]]''. Transformation is often used as a [[biotechnology]] to insert novel genes into bacteria for scientific research experiments, or for industrial or medical applications. See also [[molecular biology]] and [[biotechnology]]. | * '''Bacterial [[Transformation (genetics)|transformation]]''' , the genetic alteration of a [[cell (biology)|cell]] resulting from the introduction, uptake and [[expression (genetics)|expression]] of foreign genetic material, generally [[DNA]] (see this [http://www.learner.org/channel/courses/biology/archive/animations/hires/a_infect3_h.html animation with audio explanation]). This process is relatively common in certain naturally transformable bacteria, such as the soil bacterium ''[[Bacillus subtilis]]''. Transformation is often used as a [[biotechnology]] to insert novel genes into bacteria for scientific research experiments, or for industrial or medical applications. See also [[molecular biology]] and [[biotechnology]]. | ||
* '''[[Transduction (genetics)|Transduction]]''', the process in which bacterial DNA is moved from one bacterium to another by a bacterial virus (a bacteriophage, commonly called a [[phage]]). | * '''[[Transduction (genetics)|Transduction]]''', the process in which bacterial DNA is moved from one bacterium to another by a bacterial virus (a bacteriophage, commonly called a [[phage]]). | ||
* '''[[Bacterial conjugation]]''', a process in which a living bacterial cell transfers genetic material through direct cell-to-cell contact (see the animation [http://www.blackwellpublishing.com/trun/artwork/Animations/conjugation/conjugation.html conjugation in ''Escherichia coli'']). | * '''[[Bacterial conjugation]]''' ( or mating), a process in which a living bacterial cell transfers genetic material through direct cell-to-cell contact (see the animation [http://www.blackwellpublishing.com/trun/artwork/Animations/conjugation/conjugation.html conjugation in ''Escherichia coli'']). | ||
==Horizontal gene transfer and the problem of antibiotic resistant pathogens== | ==Horizontal gene transfer and the problem of antibiotic resistant pathogens== | ||
Antibiotic resistance in pathogenic bacteria is a serious on ongoing medical problem <ref>Kunin, C. M. (1997). Antibiotic armageddon. Clinical Infectious Diseases 25, pages 240 –1. | Antibiotic resistance in pathogenic bacteria is a serious on ongoing medical problem.<ref>Kunin, C. M. (1997). Antibiotic armageddon. Clinical Infectious Diseases 25, pages 240 –1. | ||
* [http://content.nejm.org/cgi/content/full/337/7/491?ijkey=9c7dc30f289d350760874b6d80818d46f3eb6e54&keytype2=tf_ipsecsha Swartz, M. N. (1997). Use of antimicrobial agents and drug resistance. New England Journal of Medicine 337, pages 491 –2.] | |||
* Shales, D. M., Gerding, D. N., John, J. F., Craig, W. A., Bornstein, D. L., Duncan, R. A. et al. (1997). Society for Healthcare Epidemiology of America and Infectious Diseases Society of America Joint Committee on the prevention of antimicrobial resistance: guidelines for the prevention of antimicrobial resistance in hospitals. Infection Control and Hospital Epidemiology 18, pages 275 –91.</ref> | |||
The major factor causing antibiotic resistance is the massive use of antibiotics in medicine and agriculture, and '''horizontal transfer''' is the mechanism by which this resistance evolves and spreads. Gene transfer and [[recombination]] is documented a mechanism that can generate new antibiotic resistance traits in the | The major factor causing antibiotic resistance is the massive use of antibiotics in medicine and agriculture, and '''horizontal transfer''' is the mechanism by which this resistance evolves and spreads. Gene transfer and [[recombination]] is documented a mechanism that can generate new antibiotic resistance traits in the transformation competent bacterium ''[[Streptococcus pneumoniae]]'' .<ref>Laible G, Spratt BG, Hakenback R. Interspecies recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of ''Streptococcus pneumoniae''. Mol Microbiol 1991;5:1993-2002.</ref> | ||
When one bacterial cell acquires resistance, it can readily transfer genes conferring resistance to antibiotics to many species by [[plasmid]] or [[transposon]] transfer. Plasmids and transposons provide the means for genes to persist in new host cells, plasmids by their ability to replicate | When one bacterial cell acquires resistance, it can readily transfer genes conferring resistance to antibiotics to many species by [[plasmid]] or [[transposon]] transfer. Plasmids and transposons provide the means for genes to persist in new host cells, plasmids by their ability to replicate independently as [[replicon]]s, and transposons by providing a mechanism for insertion of genes into the main chromosome. | ||
Gut bacteria for instance appear to exchange genetic material with each other within the [[gut]] in which they live. Virulence genes can be transferred by HGT.<ref>[http://jb.asm.org/cgi/content/full/187/5/1783?view=long&pmid=15716450 Wick LM, Qi W, Lacher DW, Whittam TS. (2005) J Bacteriol 2005 Mar;187(5):1783-91. Evolution of genomic content in the stepwise emergence of ''Escherichia coli'' O157:H7.]</ref> | |||
Harmless bacteria in the gut can provide a reservoir of antibiotic resistance by participating in | Harmless bacteria in the gut can provide a reservoir of antibiotic resistance by participating in HGT <ref>Shoemaker, N. B., Vlamakis, H., Hayes, K. & Salyers, A. A. (2001) Evidence for extensive resistance gene transfer among ''Bacteroides'' spp. and among ''Bacteroides'' and other genera in the human colon. Applied and Environmental Microbiology 67, 561–568 (2001).</ref>, and antibiotic resistance is a growing public-health problem because many pathogenic bacteria, including those that cause tuberculosis, pneumonia and some types of food poisoning, are increasingly unaffected by medicines. Major sources of concern are physicians over-prescribing antibiotics, and farmers for feeding antibiotics to their animals, generating resistant bacteria in the animals. | ||
==Role of horizontal gene transfer in bacterial evolution== | ==Role of horizontal gene transfer in bacterial evolution== | ||
When [[Joshua Lederberg]] discovered gene transmission among laboratory mutants of the gut bacterium ''Escherichia coli'' strain K-12 in 1948 he initiated the modern (and some say Golden) era of bacterial genetics to which he | When [[Joshua Lederberg]] discovered gene transmission among laboratory mutants of the gut bacterium ''Escherichia coli'' strain K-12 in 1948 he initiated the modern (and some say Golden) era of bacterial genetics to which he contributed many remarkable discoveries and insights. Today, our appreciation of the natural evolution of pathogenic strains of the gut bacterium he investigated in 1948 provides a satisfying vignette of current general understanding of bacterial evolution and of the importance of horizontal gene transfer in bacterial biology. To quote directly from a 2001 review article about pathogenic gut bacteria that cause disease <ref>[http://www.jci.org/cgi/content/full/107/5/539?ijkey=c8fc03bce559dc2fe6419684a38955c32dbe51a6&keytype2=tf_ipsecsha Donnenberg, M. S., and T. S. Whittam. 2001. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic ''Escherichia coli''. ''J Clin Investig'' '''107''':539-548. (Bold emphasis added)]</ref>: | ||
<blockquote>It is a mistake to think of ''E. coli'' as a | <blockquote>It is a mistake to think of ''E. coli'' as a homogeneous species. Most genes, even those encoding conserved metabolic functions, are [[polymorphic]], with multiple [[allele]]s found among different isolates <ref>Whittam, T.S. 1996. Genetic variation and evolutionary processes in natural populations of ''Escherichia coli''. In ''Escherichia coli'' and ''Salmonella'': cellular and molecular biology. F.C. Neidhardt, editor. ASM Press. Washington, DC, USA. pages 2708–2720.</ref>. The composition of the [[genome]] of ''E. coli'' is also highly dynamic. The fully sequenced genome of the laboratory K-12 strain [used by Lederberg in 1948] , whose derivatives have served an indispensable role in the laboratories of countless scientists, shows evidence of tremendous plasticity <ref>[http://www.sciencemag.org/cgi/content/abstract/277/5331/1453?ijkey=4fddae48f66054e7260b33240490332b221e5a49&keytype2=tf_ipsecsha Blattner, F.R. et al.1997. The complete genome sequence of ''Escherichia coli'' K-12. Science. 277:1453-1462.]</ref>. It has been estimated that the K-12 lineage has experienced more than 200 lateral (horizontal) transfer events since it diverged from ''Salmonella'' about 100 million years ago and that 18% of its contemporary genes were obtained horizontally from other species <ref>[http://www.pnas.org/cgi/content/abstract/95/16/9413?ijkey=927d4d942fdc85906b6d0f1f104192398d2e6f60&keytype2=tf_ipsecsha Lawrence, J.G., and Ochman, H. 1998. Molecular archeology of the ''Escherichia coli'' genome. Proc. Natl. Acad. Sci. USA. 95:9413-9417.]</ref>. Such fluid gain and loss of genetic material are also seen in the recent comparison of the genomic sequence of a pathogenic ''E. coli'' O157:H7 with the K-12 genome. Approximately 4.1 million base pairs of 'backbone' sequences are conserved between the genomes, but these stretches are punctuated by hundreds of sequences present in one strain but not in the other. The pathogenic strain contains 1.34 million base pairs of lineage-specific DNA that includes 1,387 new genes; some of these have been implicated in virulence, but many have no known function <ref>Perna, N.T. et al.2001. Genome sequence of enterohaemorrhagic ''Escherichia coli'' O157:H7. Nature. 409:529-533.</ref>. | ||
: | : | ||
: | : | ||
| Line 44: | Line 45: | ||
The [[virulence factor]]s that distinguish the various ''E. coli'' pathotypes were acquired from numerous sources, including [[plasmid]]s, [[bacteriophage]]s, and the [[genomes]] of other bacteria. '''[[Pathogenicity island]]s''', relatively large (>10 kb) genetic elements that encode virulence factors and are found specifically in the genomes of pathogenic strains, frequently have base compositions that differ drastically from that of the content of the rest of the ''E. coli'' genome, indicating that they were acquired from another species.</blockquote> | The [[virulence factor]]s that distinguish the various ''E. coli'' pathotypes were acquired from numerous sources, including [[plasmid]]s, [[bacteriophage]]s, and the [[genomes]] of other bacteria. '''[[Pathogenicity island]]s''', relatively large (>10 kb) genetic elements that encode virulence factors and are found specifically in the genomes of pathogenic strains, frequently have base compositions that differ drastically from that of the content of the rest of the ''E. coli'' genome, indicating that they were acquired from another species.</blockquote> | ||
To get a full appreciation of how the above quoted statement could be deduced from clinical and laboratory investigations, it | To get a full appreciation of how the above quoted statement could be deduced from clinical and laboratory investigations, it is helpful to take the intellectually fruitful journey along the history of bacterial genetics. | ||
==Time line for major discoveries on horizontal gene transfer in prokaryotes== | ==Time line for major discoveries on horizontal gene transfer in prokaryotes== | ||
'''1928.''' Discovery of gene transfer in bacteria started in 1928 when Frederick Griffith found ''Diplococcus pneumoniae'' bacteria (now usually called ''Streptococcus pneumoniae'') could inherit characteristics (namely the | '''1928.''' Discovery of gene transfer in bacteria started in 1928 when Frederick Griffith found ''Diplococcus pneumoniae'' bacteria (now usually called ''Streptococcus pneumoniae'') could inherit characteristics (namely the 'Smooth' phenotype affecting the bacterial cell coat (capsule)) from non-living extracts of other bacteria. This change was termed [[transformation]]. Oswald T. Avery at the Rockefeller Institute in 1944 showed that this transforming material of Diplococcus was [[DNA]]. | ||
'''1946.''' [[Joshua Lederberg]] and [[Edward Tatum]] in a series of very carefully designed experiments managed very low-frequency of genetic transfer between ''Escherichia coli'' K-12 bacteria when they were in contact with one-another <ref>Lederberg, J. and Tatum, E. L. (1946). Novel genotypes in mixed cultures of biochemical mutants of bacteria. Cold Spring | '''1946.''' [[Joshua Lederberg]] and [[Edward Tatum]] in a series of very carefully designed experiments managed very low-frequency of genetic transfer between ''Escherichia coli'' K-12 bacteria when they were in contact with one-another <ref>Lederberg, J. and Tatum, E. L. (1946). Novel genotypes in mixed cultures of biochemical mutants of bacteria. Cold Spring Harbor Symposia of Quantitative Biology. 11, p113.</ref>, and this process was later recognized to be caused by the [[plasmid]], fertility factor F. | ||
'''1951''' Joshua and Ester Lederberg , together with Zinder and Lively reported another case of gene transfer that did not need cell-to-cell contact. In this case the bacteria were ''Salmonella'' and it was later demonstrated that a bacterial virus (bacteriophage) was | '''1951''' Joshua and Ester Lederberg , together with Zinder and Lively reported another case of gene transfer that did not need cell-to-cell contact. In this case the bacteria were ''Salmonella'' and it was later demonstrated that a bacterial virus (bacteriophage) was carrying the genes between cells, and this is now called [[transduction]]. | ||
'''1960s.''' DNA transformation was demonstrated in a wide variety of bacterial species including species of ''Streptococcus, Haemophilus, Neisseria, Bacillus'', ''Synechococcus'' [[cyanobacteria]], and ''Rhizobium''. Later studies showed that is usually a highly evolved complex biological adaptation for genetic exchange,<ref>Dubnau, D. (1999). DNA uptake in bacteria. Annual Reviews in Microbiology 53, pages 217-244.</ref> and, for instance, in ''Bacillus'' depends on inter-cellular | '''1960s.''' DNA transformation was demonstrated in a wide variety of bacterial species including species of ''Streptococcus, Haemophilus, Neisseria, Bacillus'', ''Synechococcus'' [[cyanobacteria]], and ''Rhizobium''. Later studies showed that is usually a highly evolved complex biological adaptation for genetic exchange,<ref>Dubnau, D. (1999). DNA uptake in bacteria. Annual Reviews in Microbiology 53, pages 217-244.</ref> and, for instance, in ''Bacillus'' depends on inter-cellular signaling and [[Quorum sensing]] processes and is triggered by stress. | ||
====Transmissible plasmids in microorganisms==== | ====Transmissible plasmids in microorganisms==== | ||
'''1950-1958.''' Conjugation in ''E. coli'' is shown to be a one way (F+ donor to F- recipient), process, and not two-way cell fusion | '''1950-1958.''' Conjugation in ''E. coli'' is shown to be a one way (F+ donor to F- recipient), process, and not two-way cell fusion. The ability to do this is encoded by a DNA containing factor F 250,000 base pairs in size, whose transfer converts recipients to capability of doing further DNA donation, and which can move infectiously through a recipient bacterial population. Simply, put, sex is infectious in bacteria.<ref>Hayes, W. (1970) The Genetics of Bacteria and their Viruses. 2nd Edition, Blackwell.</ref> F factor was found to be able to move by conjugation to different species such as ''Salmonella''. | ||
'''1958.''' The existence of several genetic structures that can insert within bacterial chomosomes, based on observation of the bacteriophage lambda and fertility factor in 1958 F lead | '''1958.''' The existence of several genetic structures that can insert within bacterial chomosomes, based on observation of the bacteriophage lambda and fertility factor in 1958 F, lead Francois Jacob and Elie L. Wollman <ref>Jacob, F. and Wollman, E. L. (1958) Les episomes, elements genetiques ajoutes. C. R. Acad, Sci. Paris, 247, p154. </ref> to coin the term '''episome''' for DNA elements that have alternate modes of existence within the cell, either in the chromosome, or as autonomously replicating structures. Subsequent study of these phenomenon revealed numerous occurrences of mobile DNA in a wide range of organisms <ref> Berg, D. E. and Howe, M. M. (Eds.)(1989). Mobile DNA. American Society for Microbiology. Washington, D.C.</ref>(such as the presence of '''insertion sequence (IS) 'jumping genes' (discussed further below) that allow F plasmid insertion in the chromosome) and widespread horizontal gene transfer involving by bacteriophage, plasmids and mobile DNA in general. | ||
'''1959.''' Tomoichiro Akiba and Kunitaro Ochia discover '''mobile antibiotic resistance genes''' in bacteria <ref> Ochia, K. Yamanaka, T. Kimura, K. and Sawada, O. (1959). Inheritance of drug resistance (and its transfer) between ''Shigella'' strains and between ''Shigella'' and ''E. coli'' strains. Nihon Iji Shimpo 1861: p34 (In Japanese)</ref>, and the horizontal transfer is later shown to mediated by [[plasmids]] that inject DNA promiscuously into other cells <ref> S. Falcow (1975)Infectious Multiple Drug Resistance. Pion Press, London.</ref>. | '''1959.''' Tomoichiro Akiba and Kunitaro Ochia discover '''mobile antibiotic resistance genes''' in bacteria <ref> Ochia, K. Yamanaka, T. Kimura, K. and Sawada, O. (1959). Inheritance of drug resistance (and its transfer) between ''Shigella'' strains and between ''Shigella'' and ''E. coli'' strains. Nihon Iji Shimpo 1861: p34 (In Japanese)</ref>, and the horizontal transfer is later shown to mediated by [[plasmids]] that inject DNA promiscuously into other cells <ref> S. Falcow (1975)Infectious Multiple Drug Resistance. Pion Press, London.</ref>. | ||
'''1964.''' Brinton establishes that F factor containing bacteria have [[pili]] ( | '''1964.''' Brinton establishes that F factor containing bacteria have [[Pilus|pili]] (fimbriae) fiber-like appendages that are involved making contact with recipients to they can transfer DNA. Similar pili are specified by some R-plasmids (see [[Pilus]]). | ||
'''1960s.''' R-plasmids are established to be promiscuously transferred among different enteric bacteria. Typically plasmids can be transferred easily among the different species of common gut Gram negative bacteria. Transmissible plasmids are common in stool bacteria, and typically 10-20% of gut ''E. coli'' of healthy people (1960-1970) have R- | '''1960s.''' R-plasmids are established to be promiscuously transferred among different enteric bacteria. Typically plasmids can be transferred easily among the different species of common gut Gram negative bacteria. Transmissible plasmids are common in stool bacteria, and typically 10-20% of gut ''E. coli'' of healthy people (1960-1970) have R-plasmids and are antibiotic resistant<ref> Falcow, S. (1975). Infectious multiple Drug Resistance, Pion. </ref>. Bacteria are discovered to produce colicins, lethal compounds that kill other bacteria which are specified by transmissible plasmids similar to F plasmid. Examples are ColE, ColV plasmids. | ||
It is also fully | It is also fully realized that F-factor is not unique, but a special case of a very general phenomenon in bacteria of transmissible and mobile DNA, including R-plasmids, colicin determining (Col) plasmids, and prophages. Interactions between them were discovered, for instance R-plasmids were found which interfered with F-fertility. Plasmids were also discovered that carried bacterial virulence genes, Hemolysin (Hly), and surface antigens (K88). Plasmids are shown to be closed circular DNA molecules which are much smaller than the generally circular bacterial genome. | ||
====Insertion sequences (IS), transposons (Tn) and Mu==== | ====Insertion sequences (IS), transposons (Tn) and Mu==== | ||
'''1968.''' James Shapiro discovers that spontaneously | '''1968.''' James Shapiro discovers that spontaneously occurring insertions of large inserts of extra DNA can causes mutations in the galactose genes of the bacterium ''Escherichia coli'' <ref> Shapiro, J. (1969) Mutations caused by the insertion of genetic material into the galactose operon of ''Escherichia coli''. J. Molec. Biol. 40, p93-109.</ref>. This discovery ultimately led to the discovery of mobile '''[[insertion sequences]] (IS)'''. Their similarity to [[Barbara McClintock]]'s maize transposons was immediately realized when '''IS''' were found, through electron microscopy DNA [[heteroduplex]] formation between DNA molecules from different sources and other methods for detecting [[DNA hybridization]] to be natural residents of the ''E. coli'' chromosome, and frequently components of natural plasmids <ref> Galas, D. J. and Chandler, M. (1989). Bacterial insertion sequences. In Mobile DNA Berg, D E and Howe, E, eds. (Washington, DC: American Society for Microbiology), pp. 109-162.</ref>. F-factor carries more than one '''IS''' element, and they (as '''Tn1000''') are implicated in enabling bacterial chromosomes to take part in F-mediated conjugation. | ||
'''1963-1995.''' Mutator phage Mu of ''E. coli'' which inserts at numerous different sites in the ''E. coli'' chromosomes as a [[prophage]], and replicates vegetatively by repeated [[transposition]], provides a convenient tool for elucidation of the mechanisms of transposition in great detail <ref>Mizuuchi, K. (1992) Transpositional recombination: mechanistic insights from studies of Mu and other elements. Annu. Rev. Biochem. 61, 1011-1051.</ref> <ref>Pato, M. L. (1989) Bacteriophage Mu. In Mobile DNA Berg, D E and Howe, E, eds. (Washington, DC: American Society for Microbiology), pp. 23-52.</ref>. Transposition is usually a rare event, | '''1963-1995.''' Mutator phage Mu of ''E. coli'' which inserts at numerous different sites in the ''E. coli'' chromosomes as a [[prophage]], and replicates vegetatively by repeated [[transposition]], provides a convenient tool for elucidation of the mechanisms of transposition in great detail <ref>Mizuuchi, K. (1992) Transpositional recombination: mechanistic insights from studies of Mu and other elements. Annu. Rev. Biochem. 61, 1011-1051.</ref> <ref>Pato, M. L. (1989) Bacteriophage Mu. In Mobile DNA Berg, D E and Howe, E, eds. (Washington, DC: American Society for Microbiology), pp. 23-52.</ref>. Transposition is usually a rare event, and is difficult to analyze biochemically with most [[transposon]]s and [[insertion sequences]]. The more frequent "jumping gene" activity of Mu phage facilitated its elucidation as an understandable chemical reaction. | ||
'''1974.''' The first transposition of a gene for ampicillin resistance from one R-plasmid to another plasmid is fully | '''1974.''' The first transposition of a gene for ampicillin resistance from one R-plasmid to another plasmid is fully characterized <ref>Hedges, R. W. and Jacob, A. F. Transposition of ampicillin resistance from RP4 to other [[replicon]]s. Mol. Gen. Genetics 132 pages 31-40. </ref>. The ampicillin resistance [[transposon]] is named '''Tn3'''. | ||
Other transposons are '''Tn5''' (kanamycin resistance with IS50 at each termini) , and '''Tn10''' ( | Other transposons are '''Tn5''' (kanamycin resistance with IS50 at each termini) , and '''Tn10''' ( tetracycline resistance). R-plasmids are currently thought of as modular [[replicon]]s put together with '''Tn'''s and '''replication origins''' ('''ori''' sites ) providing plasmid-specific sites for initiation of [[DNA replication]], and transposition of '''Tn'''s is seen to be part of plasmid natural evolution. | ||
====Backbones and islands found in genomes==== | ====Backbones and islands found in genomes==== | ||
'''1990-present.''' Availability of many [[genome]] DNA | '''1990-present.''' Availability of many [[genome]] DNA sequences reveals bacteria such as ''E. coli'' have genomes which consist of a "backbone" of conserved housekeeping genes interrupted by "islands" of changeable genes that are foreign DNA derived inserts (such as inserted [[prophages]]) gained during stepwise evolution by gene addition<ref>[http://jb.asm.org/cgi/content/full/187/5/1783?view=long&pmid=15716450 Wick LM, Qi W, Lacher DW, Whittam TS. (2005) J Bacteriol 2005 Mar;187(5):1783-91. Evolution of genomic content in the stepwise emergence of ''Escherichia coli'' O157:H7.]</ref> . Lawrence and Ochman (1998) <ref>[http://www.pnas.org/cgi/content/abstract/95/16/9413?ijkey=927d4d942fdc85906b6d0f1f104192398d2e6f60&keytype2=tf_ipsecsha Lawrence, J.G., and Ochman, H. 1998. Molecular archaeology of the ''Escherichia coli'' genome. Proc. Natl. Acad. Sci. USA. 95:9413-9417.]</ref> calculate there is constant turnover of horizontally transferred DNA in these islands in ''E. coli'' over evolutionary time at the rate of ~16 thousand base-pairs per million years. | ||
'''2001.''' The team of Perna and others who performed comprehensive genome analysis of the the ''E. coli'' HUS pathogen in 2001 found that with this pathogen - enterohaemorrhagic ''[[Escherichia coli O157:H7]]'' - lateral gene transfer is far more extensive than ever previously previously expected. In fact, 1,387 new genes encoded in strain-specific clusters of different sizes were found in O157:H7. These include candidate virulence factors, alternative metabolic capacities, and several prophages and other new functions <ref>Perna NT, Plunkett G 3rd, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001 Jan 25;409(6819):529-33.</ref>. | '''2001.''' The team of Perna and others who performed comprehensive genome analysis of the the ''E. coli'' HUS pathogen in 2001 found that with this pathogen - enterohaemorrhagic ''[[Escherichia coli O157:H7]]'' - lateral gene transfer is far more extensive than ever previously previously expected. In fact, 1,387 new genes encoded in strain-specific clusters of different sizes were found in O157:H7. These include candidate virulence factors, alternative metabolic capacities, and several prophages and other new functions <ref>Perna NT, Plunkett G 3rd, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001 Jan 25;409(6819):529-33.</ref>. | ||
The genomic islands of bacteria associated with virulence get the special name "[[pathogenicity island]]s" (PAI). Studies of pathogenic bacteria ''E. coli'' O157:H7 indicate these have very many genes in islands, including the complex and interesting LEE (locus of enterocyte effacement) pathogenicity island <ref> Gal-Mor, O. and Finlay, B. B. (2006) Pathogenicity islands: a molecular toolbox for bacterial virulence. Cellular Microbiology 8:11, 1707-1719 </ref>; [[tRNA]] genes are frequently found at the junctions of pathogenicity islands, and these are conserved sites recognised by many prophages as targets for insertion of prophage DNA in bacterial genomes. Pathogenicity islands have been | The genomic islands of bacteria associated with virulence get the special name "[[pathogenicity island]]s" (PAI). Studies of pathogenic bacteria ''E. coli'' O157:H7 indicate these have very many genes in islands, including the complex and interesting LEE (locus of enterocyte effacement) pathogenicity island <ref> Gal-Mor, O. and Finlay, B. B. (2006) Pathogenicity islands: a molecular toolbox for bacterial virulence. Cellular Microbiology 8:11, 1707-1719 </ref>; [[tRNA]] genes are frequently found at the junctions of pathogenicity islands, and these are conserved sites recognised by many prophages as targets for insertion of prophage DNA in bacterial genomes. Pathogenicity islands have been characterized in ''Salmonella'', ''Shigella'', ''Yersinia'', ''Helicobacter pylori'' (Cag), ''Listeria monocytogenes'' (''hly'', hemolysin ) ''Staphylococcus aureus'' (''tst'', toxic shock toxin ; Mec methicillin resistance ), and ''Clostridium difficile'' (''tcdA/B'') bacteria. | ||
'''1990s-present.''' Level of insight about evolution and role of gene transfer first achieved with ''E. coli'' are repeated in numerous other pathogenic and non-pathogenic bacteria, and in the [[Archaea]]. | '''1990s-present.''' Level of insight about evolution and role of gene transfer first achieved with ''E. coli'' are repeated in numerous other pathogenic and non-pathogenic bacteria, and in the [[Archaea]]. | ||
| Line 87: | Line 88: | ||
====Puzzles in the tree of life==== | ====Puzzles in the tree of life==== | ||
'''1990s.''' Genomic studies suggest that the past | '''1990s.''' Genomic studies suggest that the past occurrence of horizontal gene transfer obscures interpretation of early events in evolution of cells and the exact nature of the '''Universal Common Ancestor''' of cellular life <ref> Gupta, R. S. and Golding, G. B. (1996). The origin of the eukaryotic cell. Trends in Biochem Sci. '''21''':166-171</ref><ref> Brown,J.R. and Doolittle,W. F. (1997) ''Archaea'' and the prokaryote to eukaryotes transition. Microbiol and Molec Biol Reviews, 61, pages 456-502. </ref>. {{see also|Three-domain system|Tree of life}} | ||
[[Image:Campylobacter.jpg|left|frame|''Campylobacter'' cells from ''Multiple Campylobacter Genomes Sequenced''. PLoS Biol 3(1): e40 DOI: 10.1371/journal.pbio.0030040 [http://journals.plos.org/plosbiology/license.php Under Open Access License]]] | [[Image:Campylobacter.jpg|left|frame|''Campylobacter'' cells from ''Multiple Campylobacter Genomes Sequenced''. PLoS Biol 3(1): e40 DOI: 10.1371/journal.pbio.0030040 [http://journals.plos.org/plosbiology/license.php Under Open Access License]]] | ||
| Line 96: | Line 95: | ||
====Multiple Campylobacter genomes sequenced==== | ====Multiple Campylobacter genomes sequenced==== | ||
[[Image:PLoS_biology_234x60.GIF|right]] | |||
<blockquote>In 1995, the first complete bacterial genome sequence was published. Now, nearly 200 bacterial genomes have been completed, and a new one hits the scientific press most weeks. This burgeoning industry is not just scientific “stamp collecting,” however. Having all these genome sequences may provide useful clues about why some bacteria cause human disease, how to control their spread, and how to treat the infections caused by them. By comparing genome sequences, scientists can learn much more about what makes a bacteria tick than they can learn from a single sequence. | <blockquote>In 1995, the first complete bacterial genome sequence was published. Now, nearly 200 bacterial genomes have been completed, and a new one hits the scientific press most weeks. This burgeoning industry is not just scientific “stamp collecting,” however. Having all these genome sequences may provide useful clues about why some bacteria cause human disease, how to control their spread, and how to treat the infections caused by them. By comparing genome sequences, scientists can learn much more about what makes a bacteria tick than they can learn from a single sequence. | ||
| Line 118: | Line 117: | ||
==References== | ==References== | ||
{{reflist|2}}[[Category:Suggestion Bot Tag]] | |||
[[Category: | |||
Latest revision as of 06:01, 29 August 2024

In the years 1987-1996 in Vietnam there was a medically alarming appearance of resistance to the antibiotic chloramphenicol in dangerous meningococci bacteria that cause infectious meningitis, hampering the treatment of the disease in that country. Subsequent laboratory investigations revealed that the gene conferring chloramphenicol resistance on pathogenic meningococcus bacteria was identical to a previously identified mobile gene named Tn4451 found in the completely different bacteria known as Clostridium perfringens.[2] Meningococci are Gram negative and aerobic microbes, while Clostridium is Gram positive and anaerobic- about as different from one-another as bacteria can be.
This episode is but one illustration of the extensive movement of genes that takes place between distantly related microbes. This process has the technical name of horizontal gene transfer (HGT) and is defined as movement of genes between different species, or across broad taxonomic categories.
As indicated by the opening example, HGT is common among both the two major taxonomic subdivisions of bacteria-like organisms - Bacteria and Archaea - and occurs even between very distantly-related microbes. Both the Bacteria and Archaea taxonomic Domains of microorganism lack distinct nuclear cellular compartments for their DNA, and are thus classified as prokaryotes, meaning they possess a primitive nucleus, in contrast to eukaryote cells which possess well-formed nuclei. (HGT also occurs in eukaryotic protist microorganisms, and in both pathogenic and non-pathogenic microorganisms, so overall is a major feature of natural microbial evolution.)
Detailed and comprehensive DNA sequence analysis of many prokaryotic cell genomes has shown that these genomes generally consist of a conserved backbone of mainly housekeeping genes that is subject to interruption by islands of DNA. These DNA islands can change relatively frequently during prokaryote evolution due to insertion, deletion and replacement events involving by foreign DNA.[3]
Studies of antibiotic resistance genes provide convincing evidence for wide transmissibility of genes between taxonomically diverse microbial species.
Main mechanisms of gene transfer in prokaryotes
HGT between distant microbial taxa is mediated by bacteriophages, plasmids, transposons, and specialized cellular DNA uptake mechanisms. These mechanisms usually categorized with the following commonly used technical terms:
- Bacterial transformation , the genetic alteration of a cell resulting from the introduction, uptake and expression of foreign genetic material, generally DNA (see this animation with audio explanation). This process is relatively common in certain naturally transformable bacteria, such as the soil bacterium Bacillus subtilis. Transformation is often used as a biotechnology to insert novel genes into bacteria for scientific research experiments, or for industrial or medical applications. See also molecular biology and biotechnology.
- Transduction, the process in which bacterial DNA is moved from one bacterium to another by a bacterial virus (a bacteriophage, commonly called a phage).
- Bacterial conjugation ( or mating), a process in which a living bacterial cell transfers genetic material through direct cell-to-cell contact (see the animation conjugation in Escherichia coli).
Horizontal gene transfer and the problem of antibiotic resistant pathogens
Antibiotic resistance in pathogenic bacteria is a serious on ongoing medical problem.[4]
The major factor causing antibiotic resistance is the massive use of antibiotics in medicine and agriculture, and horizontal transfer is the mechanism by which this resistance evolves and spreads. Gene transfer and recombination is documented a mechanism that can generate new antibiotic resistance traits in the transformation competent bacterium Streptococcus pneumoniae .[5]
When one bacterial cell acquires resistance, it can readily transfer genes conferring resistance to antibiotics to many species by plasmid or transposon transfer. Plasmids and transposons provide the means for genes to persist in new host cells, plasmids by their ability to replicate independently as replicons, and transposons by providing a mechanism for insertion of genes into the main chromosome.
Gut bacteria for instance appear to exchange genetic material with each other within the gut in which they live. Virulence genes can be transferred by HGT.[6]
Harmless bacteria in the gut can provide a reservoir of antibiotic resistance by participating in HGT [7], and antibiotic resistance is a growing public-health problem because many pathogenic bacteria, including those that cause tuberculosis, pneumonia and some types of food poisoning, are increasingly unaffected by medicines. Major sources of concern are physicians over-prescribing antibiotics, and farmers for feeding antibiotics to their animals, generating resistant bacteria in the animals.
Role of horizontal gene transfer in bacterial evolution
When Joshua Lederberg discovered gene transmission among laboratory mutants of the gut bacterium Escherichia coli strain K-12 in 1948 he initiated the modern (and some say Golden) era of bacterial genetics to which he contributed many remarkable discoveries and insights. Today, our appreciation of the natural evolution of pathogenic strains of the gut bacterium he investigated in 1948 provides a satisfying vignette of current general understanding of bacterial evolution and of the importance of horizontal gene transfer in bacterial biology. To quote directly from a 2001 review article about pathogenic gut bacteria that cause disease [8]:
It is a mistake to think of E. coli as a homogeneous species. Most genes, even those encoding conserved metabolic functions, are polymorphic, with multiple alleles found among different isolates [9]. The composition of the genome of E. coli is also highly dynamic. The fully sequenced genome of the laboratory K-12 strain [used by Lederberg in 1948] , whose derivatives have served an indispensable role in the laboratories of countless scientists, shows evidence of tremendous plasticity [10]. It has been estimated that the K-12 lineage has experienced more than 200 lateral (horizontal) transfer events since it diverged from Salmonella about 100 million years ago and that 18% of its contemporary genes were obtained horizontally from other species [11]. Such fluid gain and loss of genetic material are also seen in the recent comparison of the genomic sequence of a pathogenic E. coli O157:H7 with the K-12 genome. Approximately 4.1 million base pairs of 'backbone' sequences are conserved between the genomes, but these stretches are punctuated by hundreds of sequences present in one strain but not in the other. The pathogenic strain contains 1.34 million base pairs of lineage-specific DNA that includes 1,387 new genes; some of these have been implicated in virulence, but many have no known function [12].
The virulence factors that distinguish the various E. coli pathotypes were acquired from numerous sources, including plasmids, bacteriophages, and the genomes of other bacteria. Pathogenicity islands, relatively large (>10 kb) genetic elements that encode virulence factors and are found specifically in the genomes of pathogenic strains, frequently have base compositions that differ drastically from that of the content of the rest of the E. coli genome, indicating that they were acquired from another species.
To get a full appreciation of how the above quoted statement could be deduced from clinical and laboratory investigations, it is helpful to take the intellectually fruitful journey along the history of bacterial genetics.
Time line for major discoveries on horizontal gene transfer in prokaryotes
1928. Discovery of gene transfer in bacteria started in 1928 when Frederick Griffith found Diplococcus pneumoniae bacteria (now usually called Streptococcus pneumoniae) could inherit characteristics (namely the 'Smooth' phenotype affecting the bacterial cell coat (capsule)) from non-living extracts of other bacteria. This change was termed transformation. Oswald T. Avery at the Rockefeller Institute in 1944 showed that this transforming material of Diplococcus was DNA.
1946. Joshua Lederberg and Edward Tatum in a series of very carefully designed experiments managed very low-frequency of genetic transfer between Escherichia coli K-12 bacteria when they were in contact with one-another [13], and this process was later recognized to be caused by the plasmid, fertility factor F.
1951 Joshua and Ester Lederberg , together with Zinder and Lively reported another case of gene transfer that did not need cell-to-cell contact. In this case the bacteria were Salmonella and it was later demonstrated that a bacterial virus (bacteriophage) was carrying the genes between cells, and this is now called transduction.
1960s. DNA transformation was demonstrated in a wide variety of bacterial species including species of Streptococcus, Haemophilus, Neisseria, Bacillus, Synechococcus cyanobacteria, and Rhizobium. Later studies showed that is usually a highly evolved complex biological adaptation for genetic exchange,[14] and, for instance, in Bacillus depends on inter-cellular signaling and Quorum sensing processes and is triggered by stress.
Transmissible plasmids in microorganisms
1950-1958. Conjugation in E. coli is shown to be a one way (F+ donor to F- recipient), process, and not two-way cell fusion. The ability to do this is encoded by a DNA containing factor F 250,000 base pairs in size, whose transfer converts recipients to capability of doing further DNA donation, and which can move infectiously through a recipient bacterial population. Simply, put, sex is infectious in bacteria.[15] F factor was found to be able to move by conjugation to different species such as Salmonella.
1958. The existence of several genetic structures that can insert within bacterial chomosomes, based on observation of the bacteriophage lambda and fertility factor in 1958 F, lead Francois Jacob and Elie L. Wollman [16] to coin the term episome for DNA elements that have alternate modes of existence within the cell, either in the chromosome, or as autonomously replicating structures. Subsequent study of these phenomenon revealed numerous occurrences of mobile DNA in a wide range of organisms [17](such as the presence of insertion sequence (IS) 'jumping genes' (discussed further below) that allow F plasmid insertion in the chromosome) and widespread horizontal gene transfer involving by bacteriophage, plasmids and mobile DNA in general.
1959. Tomoichiro Akiba and Kunitaro Ochia discover mobile antibiotic resistance genes in bacteria [18], and the horizontal transfer is later shown to mediated by plasmids that inject DNA promiscuously into other cells [19].
1964. Brinton establishes that F factor containing bacteria have pili (fimbriae) fiber-like appendages that are involved making contact with recipients to they can transfer DNA. Similar pili are specified by some R-plasmids (see Pilus).
1960s. R-plasmids are established to be promiscuously transferred among different enteric bacteria. Typically plasmids can be transferred easily among the different species of common gut Gram negative bacteria. Transmissible plasmids are common in stool bacteria, and typically 10-20% of gut E. coli of healthy people (1960-1970) have R-plasmids and are antibiotic resistant[20]. Bacteria are discovered to produce colicins, lethal compounds that kill other bacteria which are specified by transmissible plasmids similar to F plasmid. Examples are ColE, ColV plasmids.
It is also fully realized that F-factor is not unique, but a special case of a very general phenomenon in bacteria of transmissible and mobile DNA, including R-plasmids, colicin determining (Col) plasmids, and prophages. Interactions between them were discovered, for instance R-plasmids were found which interfered with F-fertility. Plasmids were also discovered that carried bacterial virulence genes, Hemolysin (Hly), and surface antigens (K88). Plasmids are shown to be closed circular DNA molecules which are much smaller than the generally circular bacterial genome.
Insertion sequences (IS), transposons (Tn) and Mu
1968. James Shapiro discovers that spontaneously occurring insertions of large inserts of extra DNA can causes mutations in the galactose genes of the bacterium Escherichia coli [21]. This discovery ultimately led to the discovery of mobile insertion sequences (IS). Their similarity to Barbara McClintock's maize transposons was immediately realized when IS were found, through electron microscopy DNA heteroduplex formation between DNA molecules from different sources and other methods for detecting DNA hybridization to be natural residents of the E. coli chromosome, and frequently components of natural plasmids [22]. F-factor carries more than one IS element, and they (as Tn1000) are implicated in enabling bacterial chromosomes to take part in F-mediated conjugation.
1963-1995. Mutator phage Mu of E. coli which inserts at numerous different sites in the E. coli chromosomes as a prophage, and replicates vegetatively by repeated transposition, provides a convenient tool for elucidation of the mechanisms of transposition in great detail [23] [24]. Transposition is usually a rare event, and is difficult to analyze biochemically with most transposons and insertion sequences. The more frequent "jumping gene" activity of Mu phage facilitated its elucidation as an understandable chemical reaction.
1974. The first transposition of a gene for ampicillin resistance from one R-plasmid to another plasmid is fully characterized [25]. The ampicillin resistance transposon is named Tn3. Other transposons are Tn5 (kanamycin resistance with IS50 at each termini) , and Tn10 ( tetracycline resistance). R-plasmids are currently thought of as modular replicons put together with Tns and replication origins (ori sites ) providing plasmid-specific sites for initiation of DNA replication, and transposition of Tns is seen to be part of plasmid natural evolution.
Backbones and islands found in genomes
1990-present. Availability of many genome DNA sequences reveals bacteria such as E. coli have genomes which consist of a "backbone" of conserved housekeeping genes interrupted by "islands" of changeable genes that are foreign DNA derived inserts (such as inserted prophages) gained during stepwise evolution by gene addition[26] . Lawrence and Ochman (1998) [27] calculate there is constant turnover of horizontally transferred DNA in these islands in E. coli over evolutionary time at the rate of ~16 thousand base-pairs per million years.
2001. The team of Perna and others who performed comprehensive genome analysis of the the E. coli HUS pathogen in 2001 found that with this pathogen - enterohaemorrhagic Escherichia coli O157:H7 - lateral gene transfer is far more extensive than ever previously previously expected. In fact, 1,387 new genes encoded in strain-specific clusters of different sizes were found in O157:H7. These include candidate virulence factors, alternative metabolic capacities, and several prophages and other new functions [28].
The genomic islands of bacteria associated with virulence get the special name "pathogenicity islands" (PAI). Studies of pathogenic bacteria E. coli O157:H7 indicate these have very many genes in islands, including the complex and interesting LEE (locus of enterocyte effacement) pathogenicity island [29]; tRNA genes are frequently found at the junctions of pathogenicity islands, and these are conserved sites recognised by many prophages as targets for insertion of prophage DNA in bacterial genomes. Pathogenicity islands have been characterized in Salmonella, Shigella, Yersinia, Helicobacter pylori (Cag), Listeria monocytogenes (hly, hemolysin ) Staphylococcus aureus (tst, toxic shock toxin ; Mec methicillin resistance ), and Clostridium difficile (tcdA/B) bacteria.
1990s-present. Level of insight about evolution and role of gene transfer first achieved with E. coli are repeated in numerous other pathogenic and non-pathogenic bacteria, and in the Archaea.
Puzzles in the tree of life
1990s. Genomic studies suggest that the past occurrence of horizontal gene transfer obscures interpretation of early events in evolution of cells and the exact nature of the Universal Common Ancestor of cellular life [30][31].
- See also: Three-domain system and Tree of life
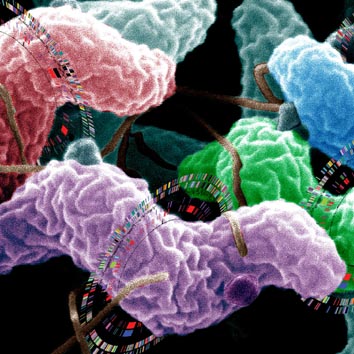
2005. A good example of how genome analysis reveals the role of horizontal gene movement in bacterial evolution and how understanding it may to better public health measures is available for Campylobacter bacteria, a cause of serious gastroenteritis [32]:
Multiple Campylobacter genomes sequenced
In 1995, the first complete bacterial genome sequence was published. Now, nearly 200 bacterial genomes have been completed, and a new one hits the scientific press most weeks. This burgeoning industry is not just scientific “stamp collecting,” however. Having all these genome sequences may provide useful clues about why some bacteria cause human disease, how to control their spread, and how to treat the infections caused by them. By comparing genome sequences, scientists can learn much more about what makes a bacteria tick than they can learn from a single sequence.
Derrick Fouts and his colleagues have taken this comparative approach with Campylobacter. Infection with a Campylobacter species is one of the most common causes of human bacterial gastroenteritis. In the US, 15 out of every 100,000 people are diagnosed with campylobacteriosis every year, and with many cases going unreported, up to 0.5% of the general population may unknowingly harbor Campylobacter in their gut annually. Diarrhea, cramps, abdominal pain, and fever develop within 2–5 days of picking up a pathogenic Campylobacter species, and in most people, the illness lasts for 7–10 days. But the infection can sometimes be fatal, and some individuals develop Guillain-Barré syndrome, in which the nerves that join the spinal cord and brain to the rest of the body are damaged, sometimes permanently.
Campylobacteriosis is usually caused by C. jejuni, a spiral-shaped bacterium normally found in cattle, swine, and birds, where it causes no problems. But the illness can also be caused by C. coli (also found in cattle, swine, and birds), C. upsaliensis (found in cats and dogs), and C. lari (present in seabirds in particular). Disease-causing bacteria generally get into people via contaminated food, often undercooked or poorly handled poultry, although contact with contaminated water, livestock, or household pets can also cause disease.
Comparison of four species of Campylobacter
In 2000, C. jejuni was the first food-borne pathogen to be completely sequenced, but we still know little about how Campylobacter species cause disease. In their search for clues, Derrick Fouts and coworkers have completely sequenced the genome of C. jejuni strain RM1221 (isolated from a chicken carcass) and compared it with the previously sequenced C. jejuni strain NCTC 11168 and with the unfinished sequences of C. coli strain RM2228 (a multi-drug-resistant chicken isolate), C. lari strain RM2100 (a clinical isolate), and C. upsaliensis strain RM3195 (taken from a patient with Guillain-Barré syndrome).
The researchers describe numerous differences and similarities between these different Campylobacter strains and species. For example, there are major structural differences between the genomes caused by the insertion of new stretches of DNA. Some of these pieces of DNA may carry genes that improve bacterial virulence or fitness, so their presence could help to explain the different biological behaviors of these strains. There are also major variations in the genes responsible for synthesis of molecules that are important for the interaction of Campylobacter with the environment. Such differences could underlie the host specificity of the different species.
Differences between the Campylobacter species in genes that are likely to be involved in aspects of bacterial virulence, such as adherence, motility, and toxin formation, are all detailed by Fouts et al., who also describe a new putative Campylobacter virulence locus. Further work is needed to relate these genomic differences to functional differences, but this detailed comparative genomic analysis provides the core blueprint for this important family of human pathogens. And in doing so, it lays the foundation for the development of new ways to monitor and control Campylobacter in the food chain and in human infection.
Citation: (2005) Multiple Campylobacter Genomes Sequenced. PLoS Biol 3(1): e40 DOI: 10.1371/journal.pbio.0030040
Published: January 4, 2005
Copyright: © 2005 Public Library of Science. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- ↑ Luria, S. E. (1947). Recent advances in bacterial genetics. Bacteriological Reviews 11, page 1.
- ↑ Galimand M, Gerbaud G, Guibourdenche M, Riou JY, Courvalin P.(1998) High-level chloramphenicol resistance in Neisseria meningitidis. N Engl J Med. 1998 Sep 24;339(13):868-74. Erratum in: Engl J Med 1999 Mar 11;340(10):824.Comment in: N Engl J Med. 1998 Sep 24;339(13):917-8.
- ↑ Levin BR Bergstrom CT (2000) Bacteria are Different: Observations, Interpretations, Speculations, and Opinions About the Mechanisms of Adaptive Evolution in Prokaryotes, Chapter 7 in Francisco J. Ayala, Walter M. Fitch, and Michael T. Clegg, Editors, Variation and Evolution in Plants and Microorganisms, Towards a New Synthesis 50 years after Stebbing. National Academy of Sciences, Washington DC. also available as Levin BR, Bergstrom CT.(2000) Bacteria are different: observations, interpretations, speculations, and opinions about the mechanisms of adaptive evolution in prokaryotes .Proc. Natl. Acad. Sci. U.S.A. 2000 Jun 20;97(13):6981-5. PMID 10860960
- ↑ Kunin, C. M. (1997). Antibiotic armageddon. Clinical Infectious Diseases 25, pages 240 –1.
- Swartz, M. N. (1997). Use of antimicrobial agents and drug resistance. New England Journal of Medicine 337, pages 491 –2.
- Shales, D. M., Gerding, D. N., John, J. F., Craig, W. A., Bornstein, D. L., Duncan, R. A. et al. (1997). Society for Healthcare Epidemiology of America and Infectious Diseases Society of America Joint Committee on the prevention of antimicrobial resistance: guidelines for the prevention of antimicrobial resistance in hospitals. Infection Control and Hospital Epidemiology 18, pages 275 –91.
- ↑ Laible G, Spratt BG, Hakenback R. Interspecies recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol 1991;5:1993-2002.
- ↑ Wick LM, Qi W, Lacher DW, Whittam TS. (2005) J Bacteriol 2005 Mar;187(5):1783-91. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7.
- ↑ Shoemaker, N. B., Vlamakis, H., Hayes, K. & Salyers, A. A. (2001) Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Applied and Environmental Microbiology 67, 561–568 (2001).
- ↑ Donnenberg, M. S., and T. S. Whittam. 2001. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J Clin Investig 107:539-548. (Bold emphasis added)
- ↑ Whittam, T.S. 1996. Genetic variation and evolutionary processes in natural populations of Escherichia coli. In Escherichia coli and Salmonella: cellular and molecular biology. F.C. Neidhardt, editor. ASM Press. Washington, DC, USA. pages 2708–2720.
- ↑ Blattner, F.R. et al.1997. The complete genome sequence of Escherichia coli K-12. Science. 277:1453-1462.
- ↑ Lawrence, J.G., and Ochman, H. 1998. Molecular archeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA. 95:9413-9417.
- ↑ Perna, N.T. et al.2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 409:529-533.
- ↑ Lederberg, J. and Tatum, E. L. (1946). Novel genotypes in mixed cultures of biochemical mutants of bacteria. Cold Spring Harbor Symposia of Quantitative Biology. 11, p113.
- ↑ Dubnau, D. (1999). DNA uptake in bacteria. Annual Reviews in Microbiology 53, pages 217-244.
- ↑ Hayes, W. (1970) The Genetics of Bacteria and their Viruses. 2nd Edition, Blackwell.
- ↑ Jacob, F. and Wollman, E. L. (1958) Les episomes, elements genetiques ajoutes. C. R. Acad, Sci. Paris, 247, p154.
- ↑ Berg, D. E. and Howe, M. M. (Eds.)(1989). Mobile DNA. American Society for Microbiology. Washington, D.C.
- ↑ Ochia, K. Yamanaka, T. Kimura, K. and Sawada, O. (1959). Inheritance of drug resistance (and its transfer) between Shigella strains and between Shigella and E. coli strains. Nihon Iji Shimpo 1861: p34 (In Japanese)
- ↑ S. Falcow (1975)Infectious Multiple Drug Resistance. Pion Press, London.
- ↑ Falcow, S. (1975). Infectious multiple Drug Resistance, Pion.
- ↑ Shapiro, J. (1969) Mutations caused by the insertion of genetic material into the galactose operon of Escherichia coli. J. Molec. Biol. 40, p93-109.
- ↑ Galas, D. J. and Chandler, M. (1989). Bacterial insertion sequences. In Mobile DNA Berg, D E and Howe, E, eds. (Washington, DC: American Society for Microbiology), pp. 109-162.
- ↑ Mizuuchi, K. (1992) Transpositional recombination: mechanistic insights from studies of Mu and other elements. Annu. Rev. Biochem. 61, 1011-1051.
- ↑ Pato, M. L. (1989) Bacteriophage Mu. In Mobile DNA Berg, D E and Howe, E, eds. (Washington, DC: American Society for Microbiology), pp. 23-52.
- ↑ Hedges, R. W. and Jacob, A. F. Transposition of ampicillin resistance from RP4 to other replicons. Mol. Gen. Genetics 132 pages 31-40.
- ↑ Wick LM, Qi W, Lacher DW, Whittam TS. (2005) J Bacteriol 2005 Mar;187(5):1783-91. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7.
- ↑ Lawrence, J.G., and Ochman, H. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA. 95:9413-9417.
- ↑ Perna NT, Plunkett G 3rd, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001 Jan 25;409(6819):529-33.
- ↑ Gal-Mor, O. and Finlay, B. B. (2006) Pathogenicity islands: a molecular toolbox for bacterial virulence. Cellular Microbiology 8:11, 1707-1719
- ↑ Gupta, R. S. and Golding, G. B. (1996). The origin of the eukaryotic cell. Trends in Biochem Sci. 21:166-171
- ↑ Brown,J.R. and Doolittle,W. F. (1997) Archaea and the prokaryote to eukaryotes transition. Microbiol and Molec Biol Reviews, 61, pages 456-502.
- ↑ Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, et al. (2005) Major Structural Differences and Novel Potential Virulence Mechanisms from the Genomes of Multiple Campylobacter Species. PLoS Biol 3(1): e15 DOI: 10.1371/journal.pbio.0030015
